Professor Sachin Goyal's research group focuses on fundamental research in core Mechanical Engineering areas of Mechanics, Dynamics or Controls motivated by applications in the fields of Biology and Medicine. The primary emphasis is on physical, conceptual and mathematical modeling along with computational simulations, and the applications range from human biomechanics to structural dynamics of micro-scale biological filaments. The quest toward such ambitious cross-disciplinary research applications in our group germinates research projects with purely engineering applications as well.
An Example Research Direction:
Modeling Constitutive Laws of Biological Filaments from their Atomistic Structures
|
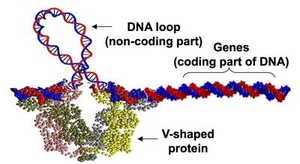
| Protein-mediated DNA looping is known to be a common gene-regulation mechanism. Experiments alone have not been able to reveal the looped structure of DNA. Continuum rod model simulations provide estimates of the loop. |
Bending and twisting of biological filaments such as DNA (Deoxyribonucleic acid) are crucial to their biological functions. For example, as shown in the Figure on the right, activity of the genes in lac-operon is governed by the sequence-dependent looping behavior of its “non-coding” DNA segment (portion of DNA that does not contain genes). Understanding the nonlinear dynamics of these mechanical deformations and how they are caused within the intra-cellular environment provides for a promising foundation for futuristic medical inventions. In fact, this ambitious and scientifically rich research quest is best introduced by posing three fundamental questions:
- How do the structural deformations of non-coding DNA regulate gene expression?
- How do the gene-regulating proteins manipulate structural deformations in DNA?
- How does base-pair sequence (or the atomistic structure) of non-coding DNA influence its structural deformability?

|
| Torsional buckling of a filament simulated from our computational rod model. For the video animation, refer to the youtube video: https://youtu.be/bGAQ-5wBYiQ. |
While several experimental biophysicists are engaged in answering the first question, our program is addressing the other two questions that ensued a rigorous and basic research in field of mechanics. Recognizing DNA as a thin filament with bending and torsional stiffness, we developed a “rod” model to simulate the biologically relevant looping and intertwining of the DNA molecules. Figure on the right shows an example simulation that involves nonlinear dynamics of torsional buckling with self-contact and intertwining. The model requires solving constrained partial differential equations that pose numerical challenges. It depicts the torsional buckling of a filament simulated from our computational rod model.
There is however a very little effort towards the third question, which actually has the grassroots of the first two questions. We are investigating the third question by analyzing how the atomic configurations and interatomic interactions map into constitutive behaviors at continuum level, i.e. into the restoring effects or “springiness” of the filament in bending and twisting. Since a first-principles derivation of the constitutive law from an atomistic-level structure and interactions is often impractical and so are direct experimental measurements due to the small length-scales, we are developing an “inverse rod model” to estimate the constitutive law from high-fidelity discrete-structure simulations such as molecular-dynamics (MD) simulations.


