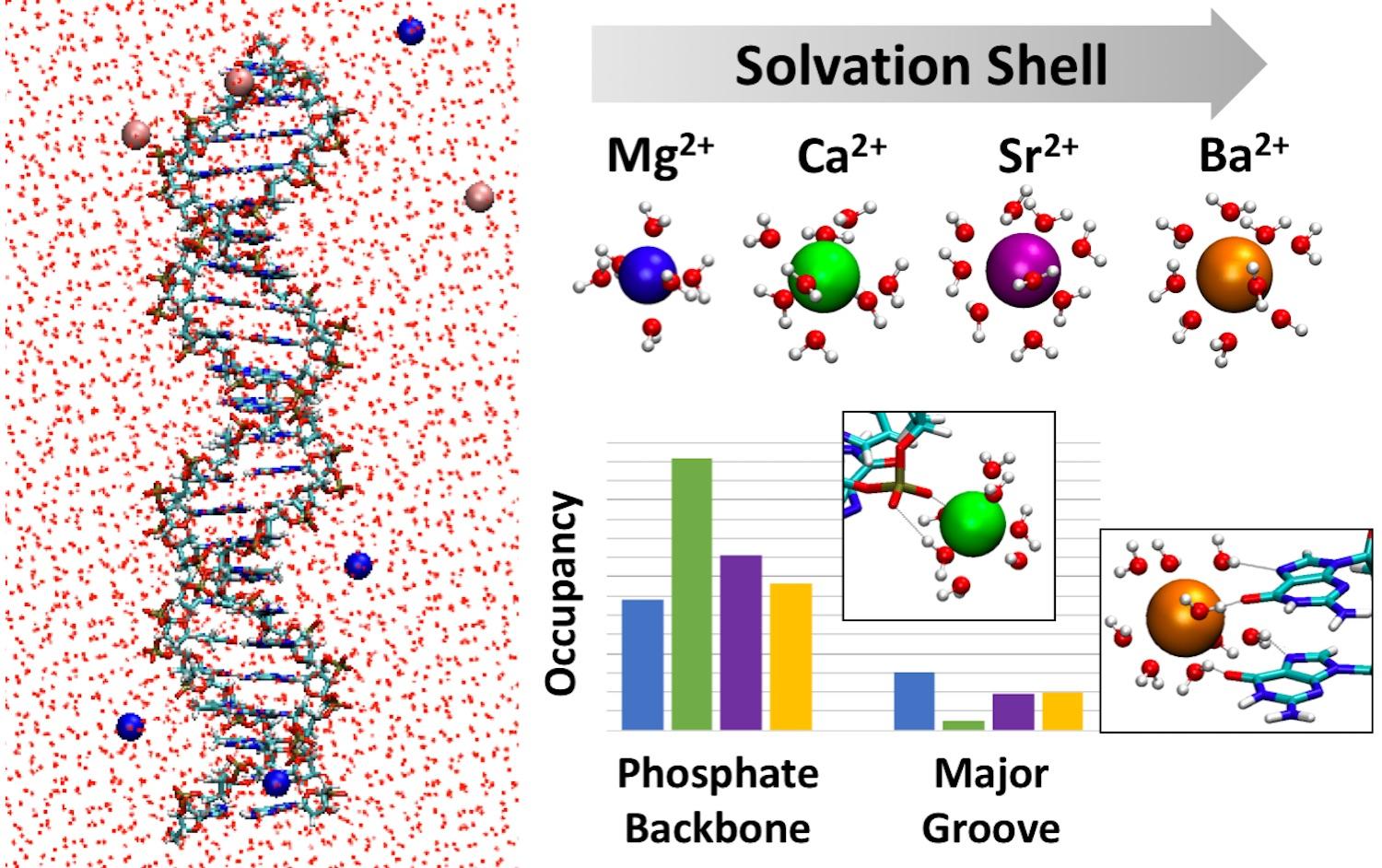
In this study published in PCCP, led by Professor Provorse Long and students at UCA, we demonstrate that the size and flexibility of the first solvation shell of alkaline earth metal ions is key to predicting trends in DNA-ion binding. We use classical atomistic molecular dynamics simulations to model double-strand DNA solvated by a series of alkaline earth metal ions (Mg2+, Ca2+, Sr2+, and Ba2+). Previous studies using similar methods have focused on small mono- and divalent ions such as K+, Na+, and Mg2+. For the first time, we expand this comparison to larger ions and reveal size-dependent trends that agree with experimental results. The atomistic detail of the simulations identify that the size and flexibility of the first solvation shell is responsible for these size-dependent trends, a point that has not been emphasized in literature thus far.
Our analysis provides a quantitative comparison of the extent to which these ions preferentially bind to specific DNA binding sites, where the size and flexibility of the first solvation shell of each ion is used to rationalize the observed trends. In addition, we find that ions bound within the major groove of dsDNA form more contacts than those bound to the phosphate backbone, regardless of ion size. These results may provide physical insight into ion-mediated interactions between dsDNA and other nanoscale structures.
This work also highlights the importance of considering the O6 atom of guanine when investigating ion binding within the major groove of dsDNA. Previous work emphasizes the N7 atom of guanine, but our work shows that the ions that bind to the major groove have a slight preference for the O6 atom rather than the N7 atom. We believe this point will inform future design and interpretation of experimental and theoretical studies of DNA-ion binding.
We believe the physical insights described in this manuscript are of interest to both experimental and theoretical physical chemists because they provide a basis for predicting DNA-ion interactions, which are key to understanding biological and material chemistry applications of DNA.
